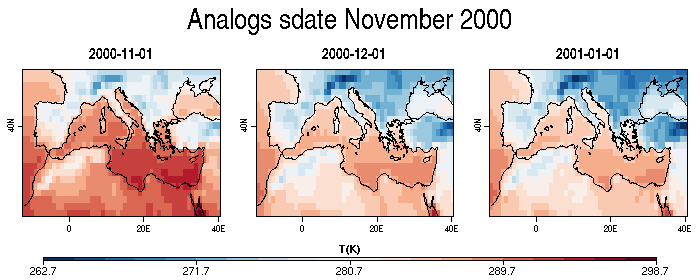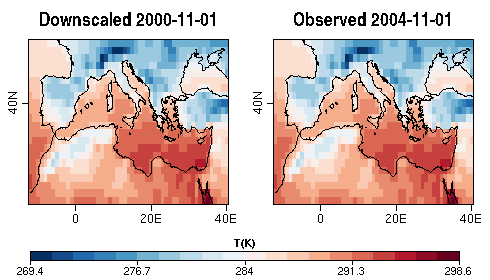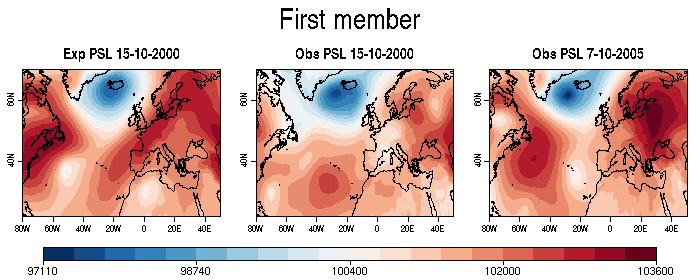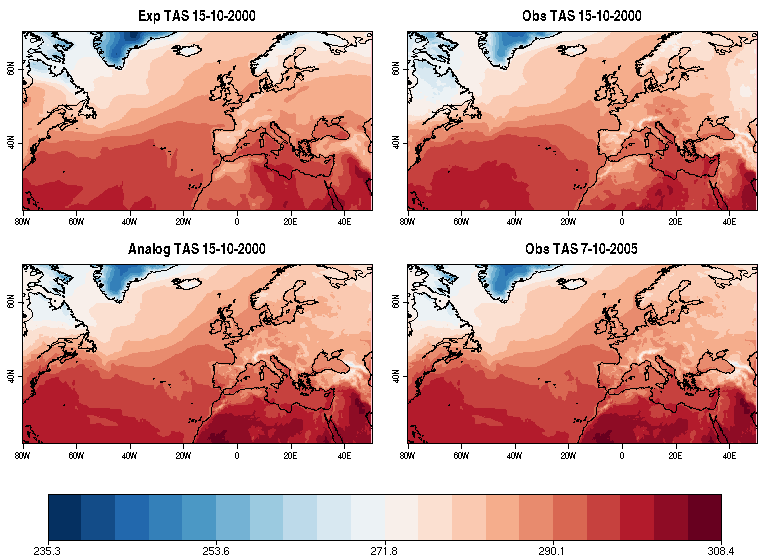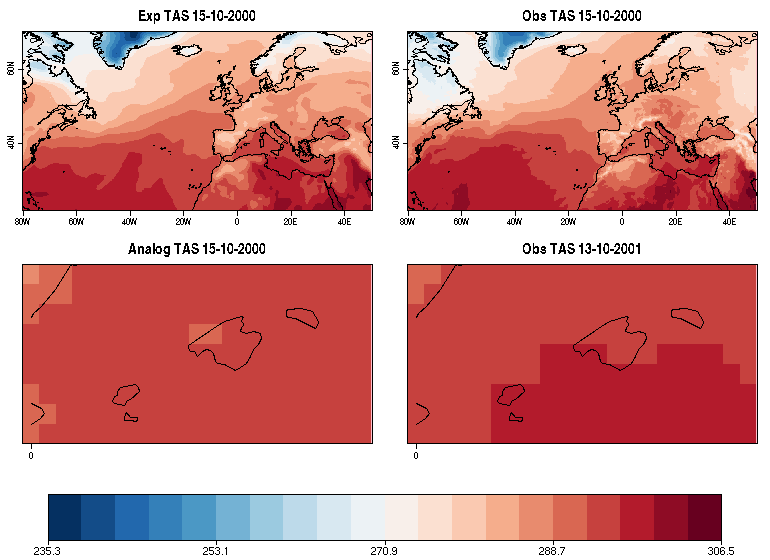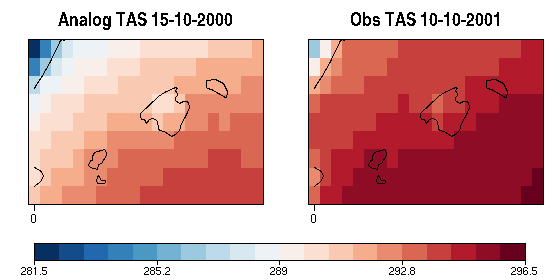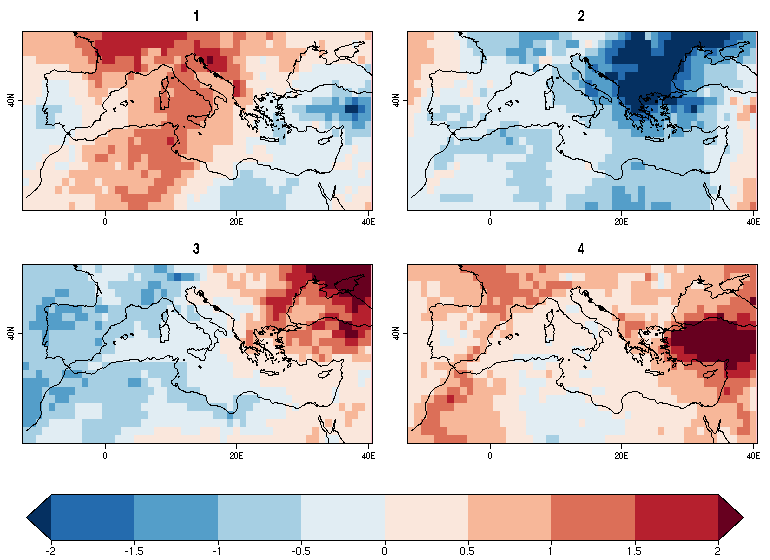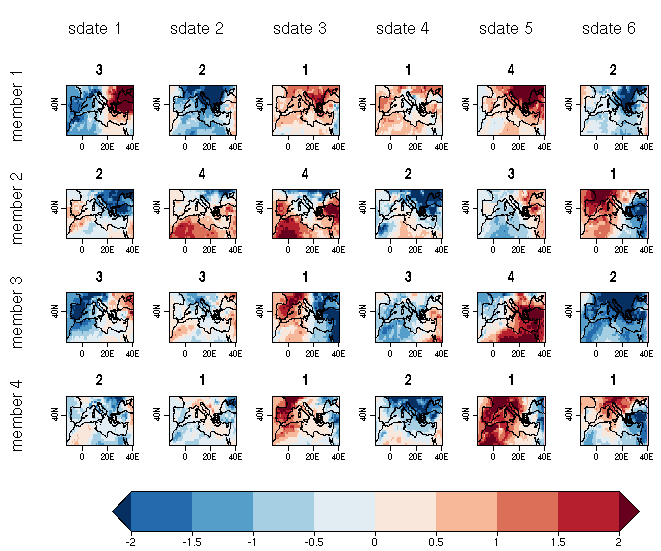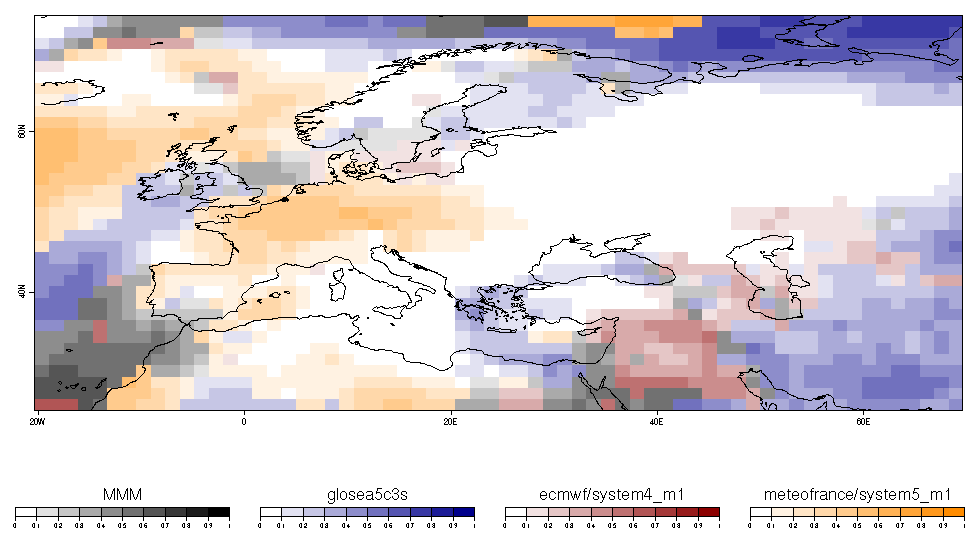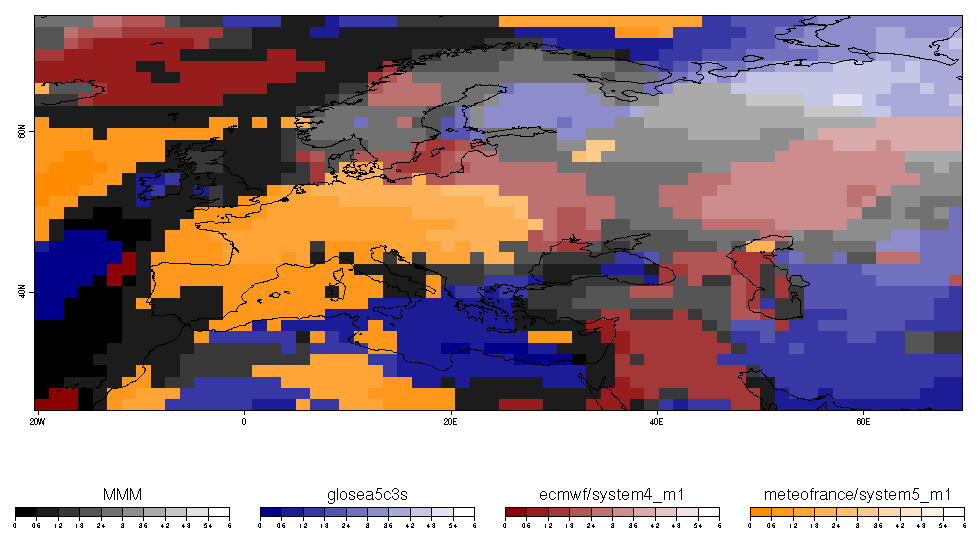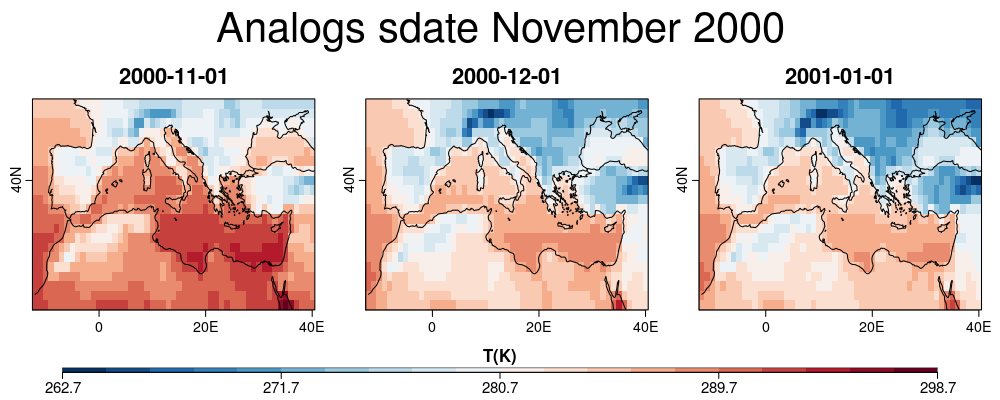
| W: | H:
| W: | H:


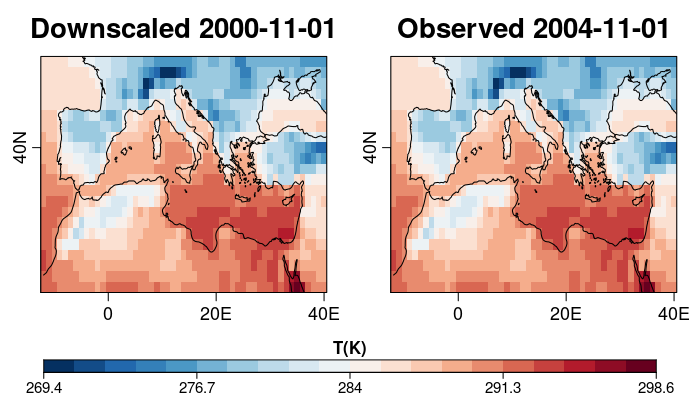
| W: | H:
| W: | H:


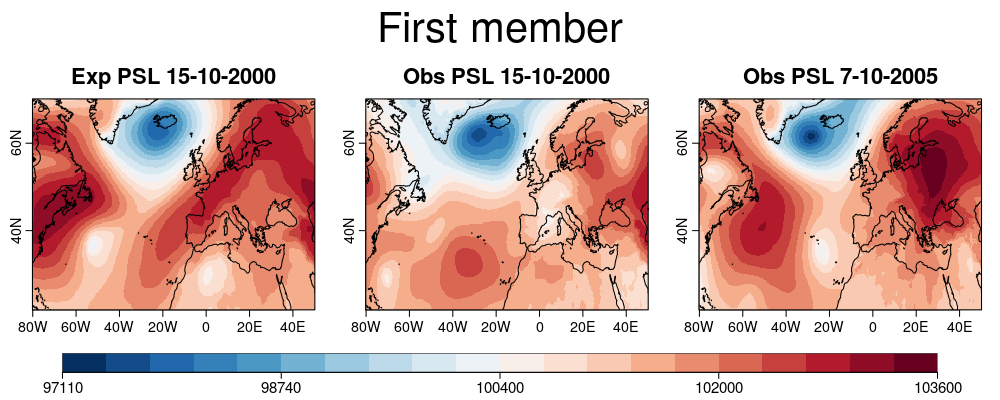
| W: | H:
| W: | H:


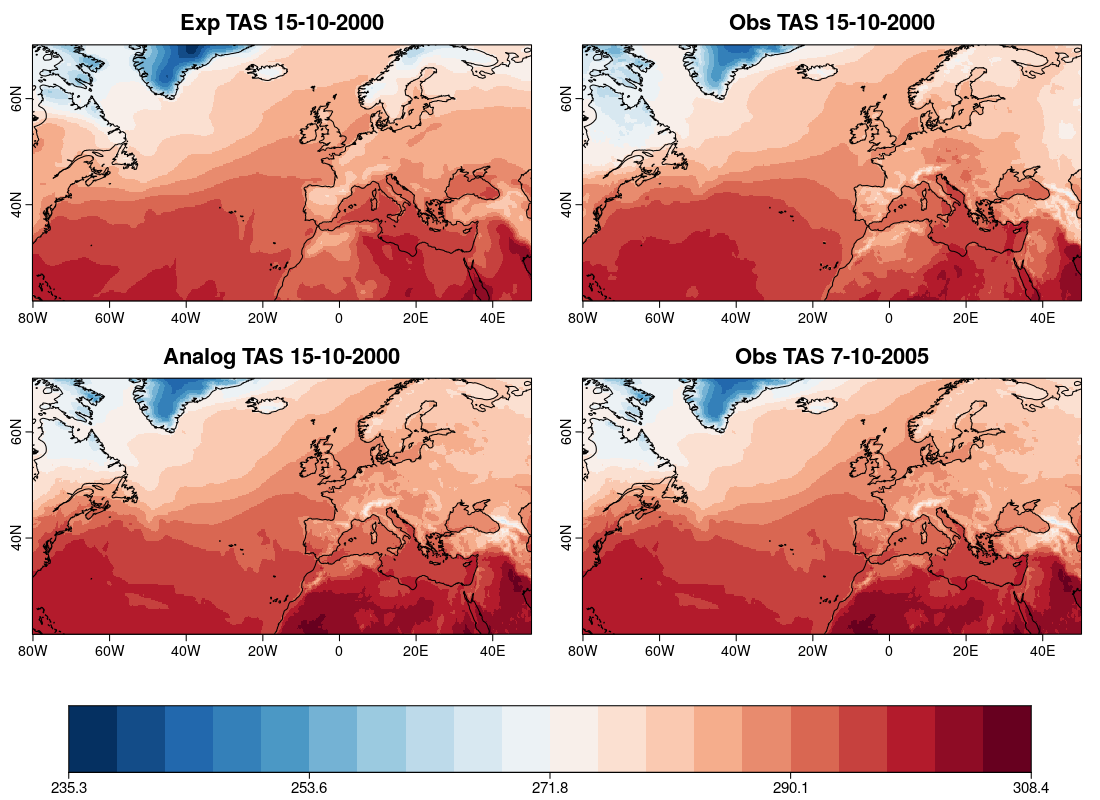
| W: | H:
| W: | H:


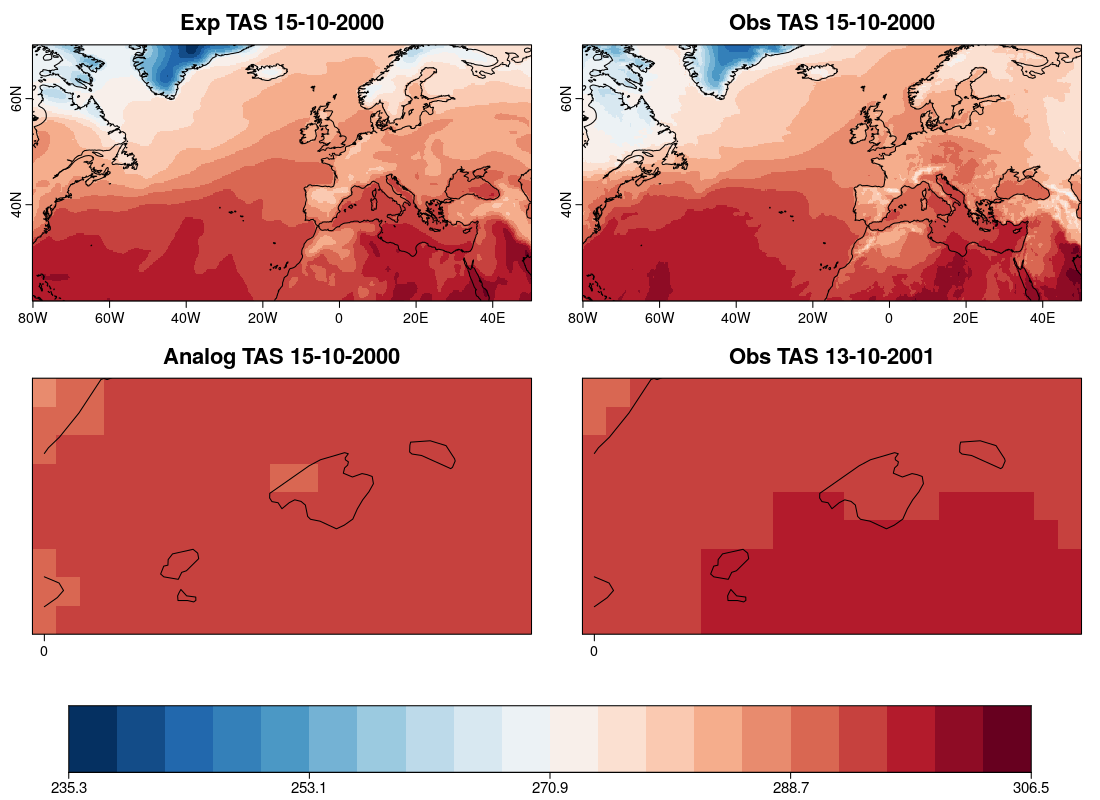
| W: | H:
| W: | H:


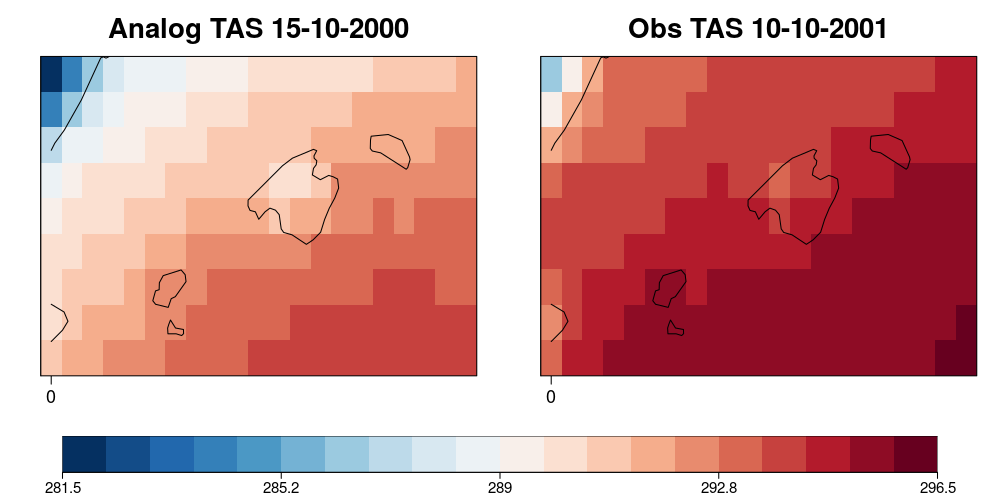
| W: | H:
| W: | H:


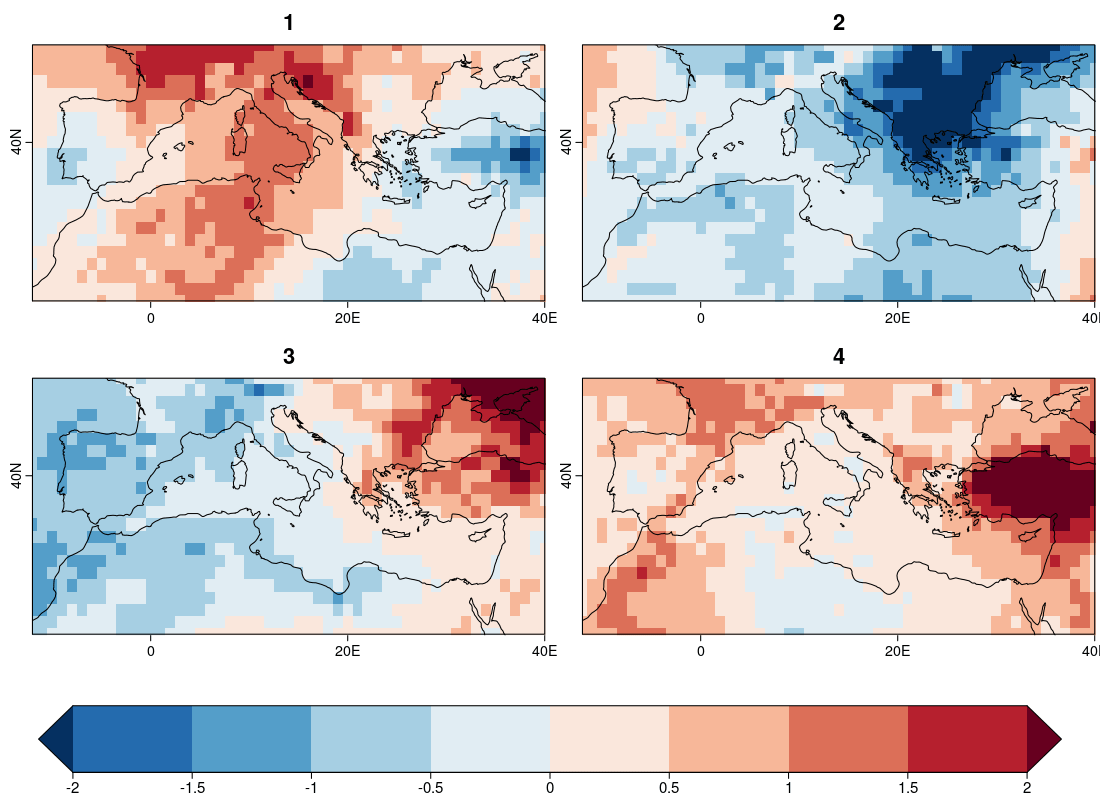
| W: | H:
| W: | H:


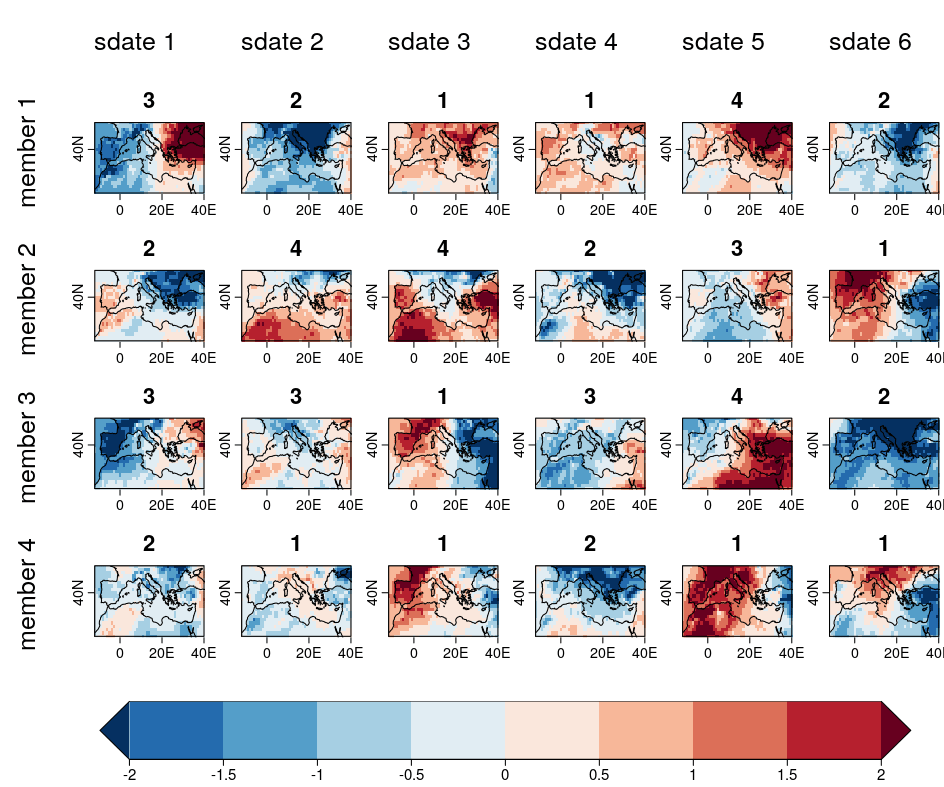
| W: | H:
| W: | H:


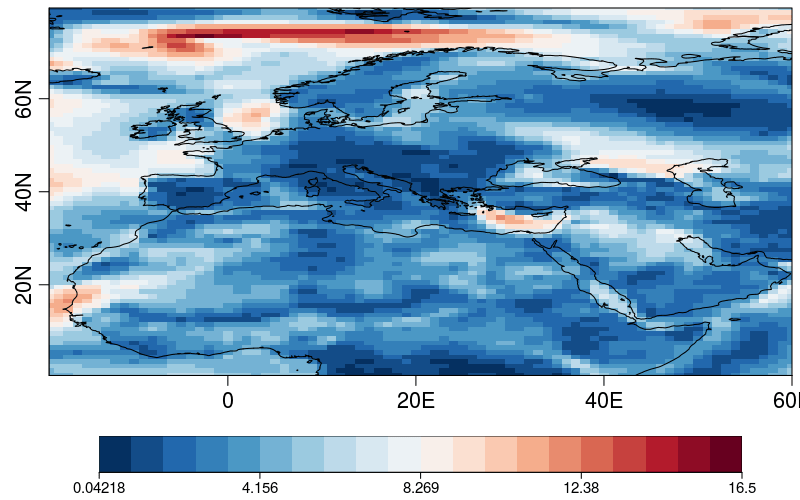
| W: | H:
| W: | H:


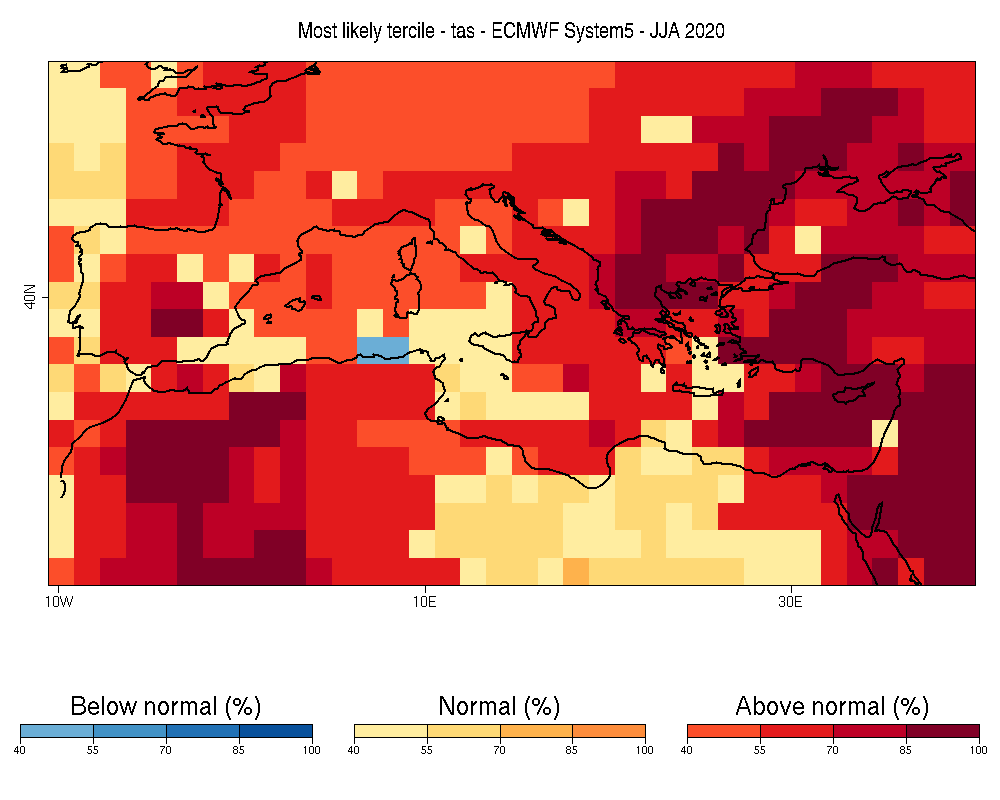
| W: | H:
| W: | H:


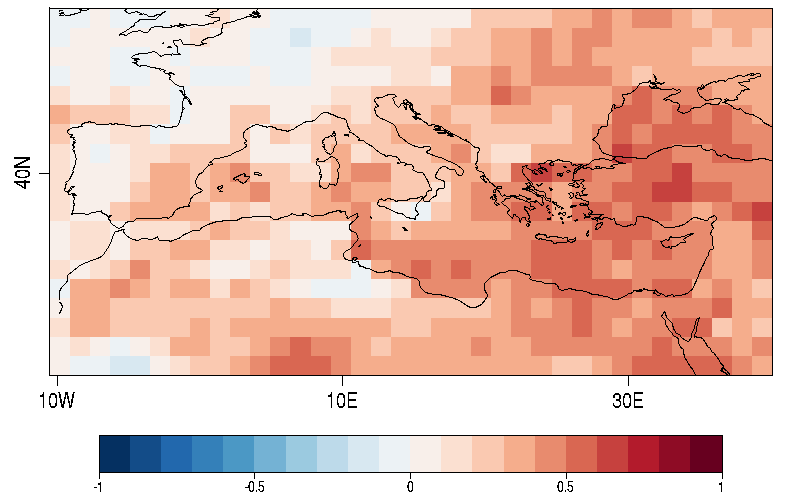
| W: | H:
| W: | H:


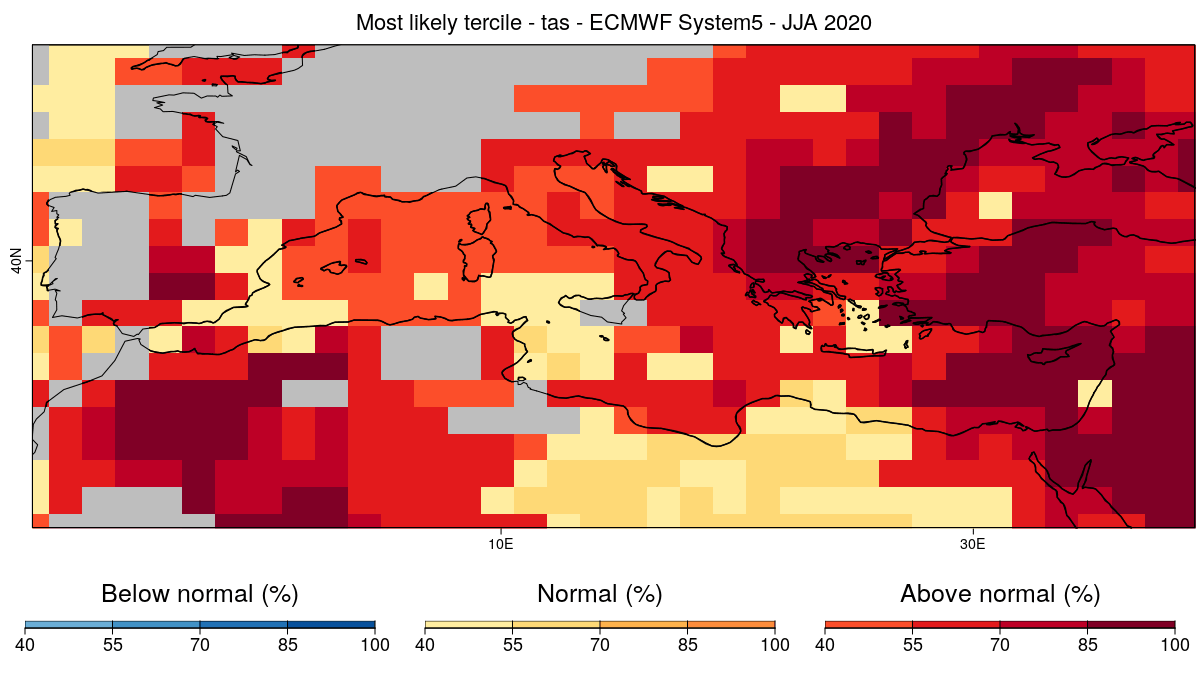
| W: | H:
| W: | H:


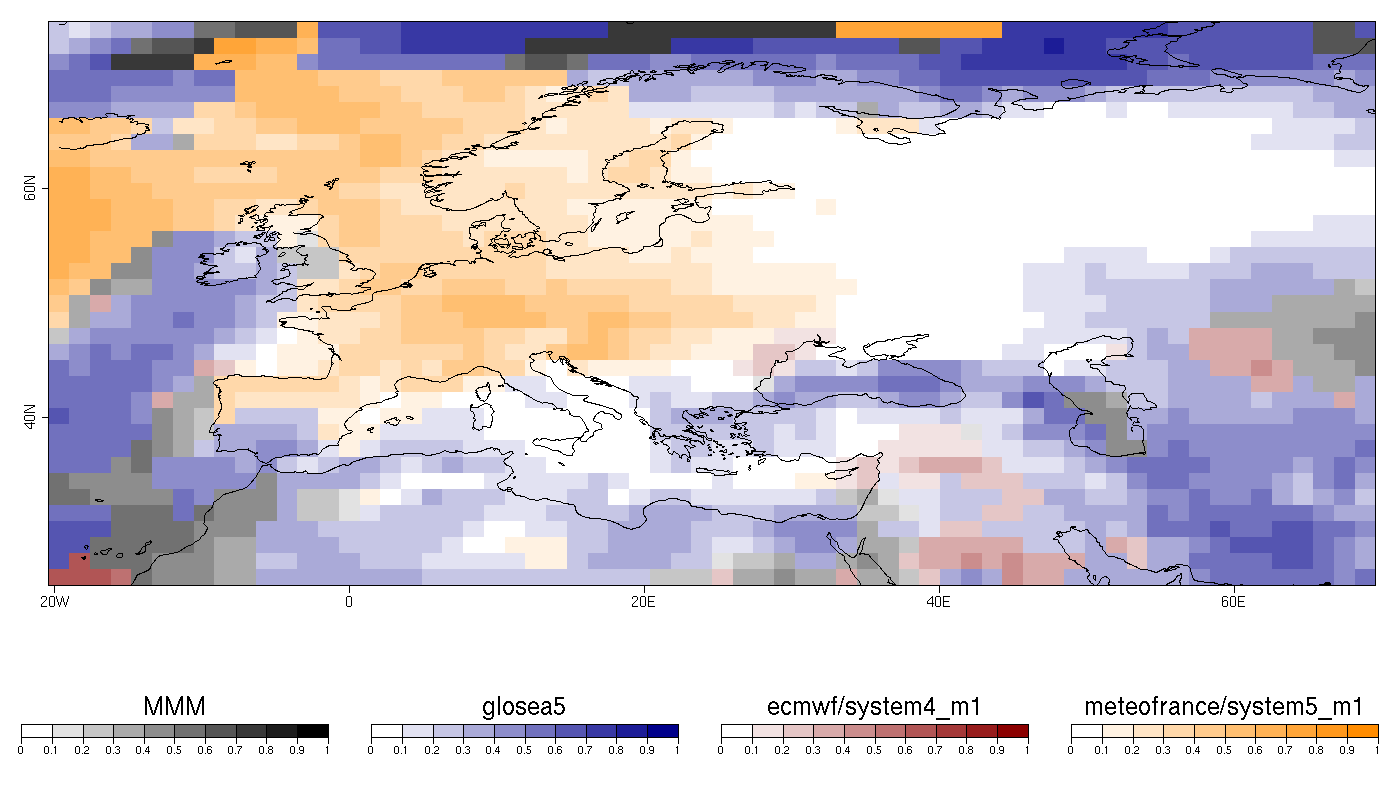
| W: | H:
| W: | H:


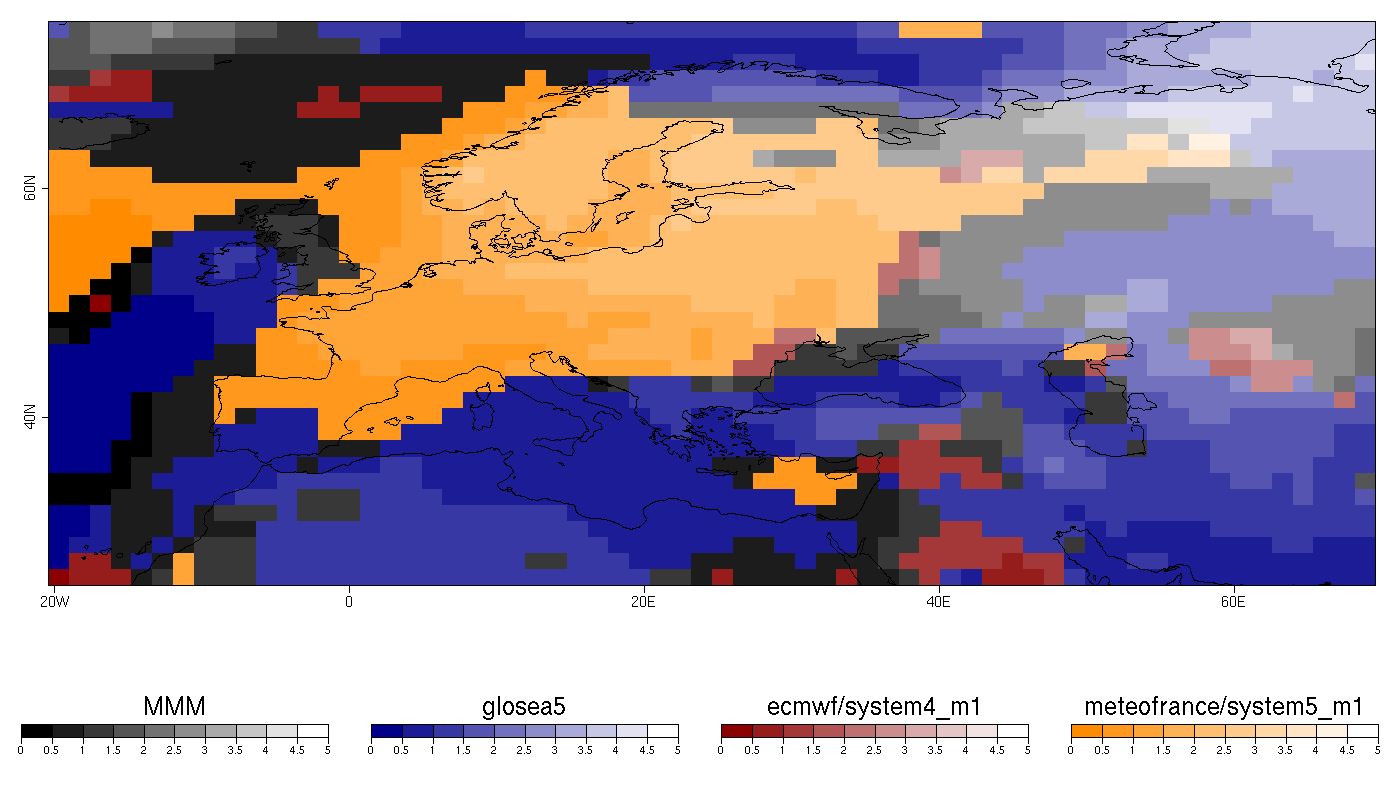
| W: | H:
| W: | H:


| W: | H:
| W: | H: