| W: | H:
| W: | H:



| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:



| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:


| W: | H:
| W: | H:



14.3 KB | W: | H:
8.99 KB | W: | H:





11.9 KB | W: | H:

24 KB | W: | H:





26.2 KB | W: | H:

26.2 KB | W: | H:





42.8 KB | W: | H:
35.8 KB | W: | H:





31.8 KB | W: | H:

28.6 KB | W: | H:





31 KB | W: | H:
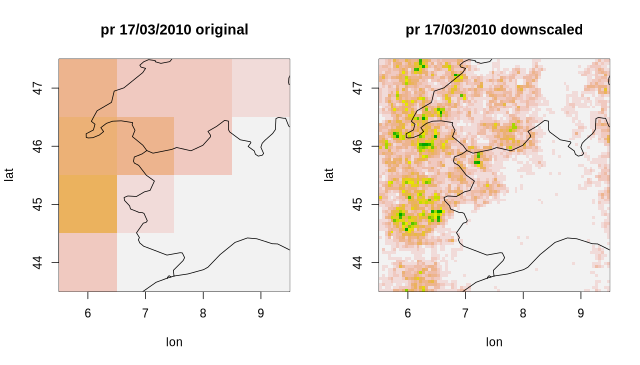
31.2 KB | W: | H:





34.9 KB | W: | H:
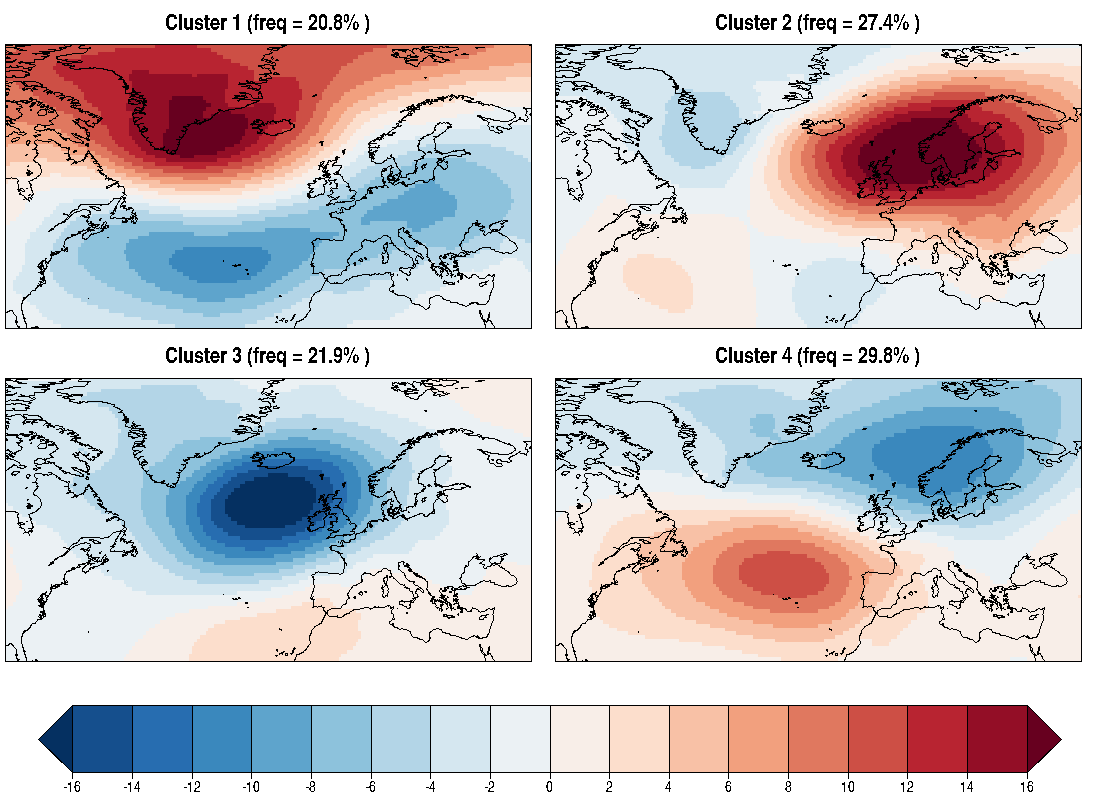
39.7 KB | W: | H:





34.1 KB | W: | H:
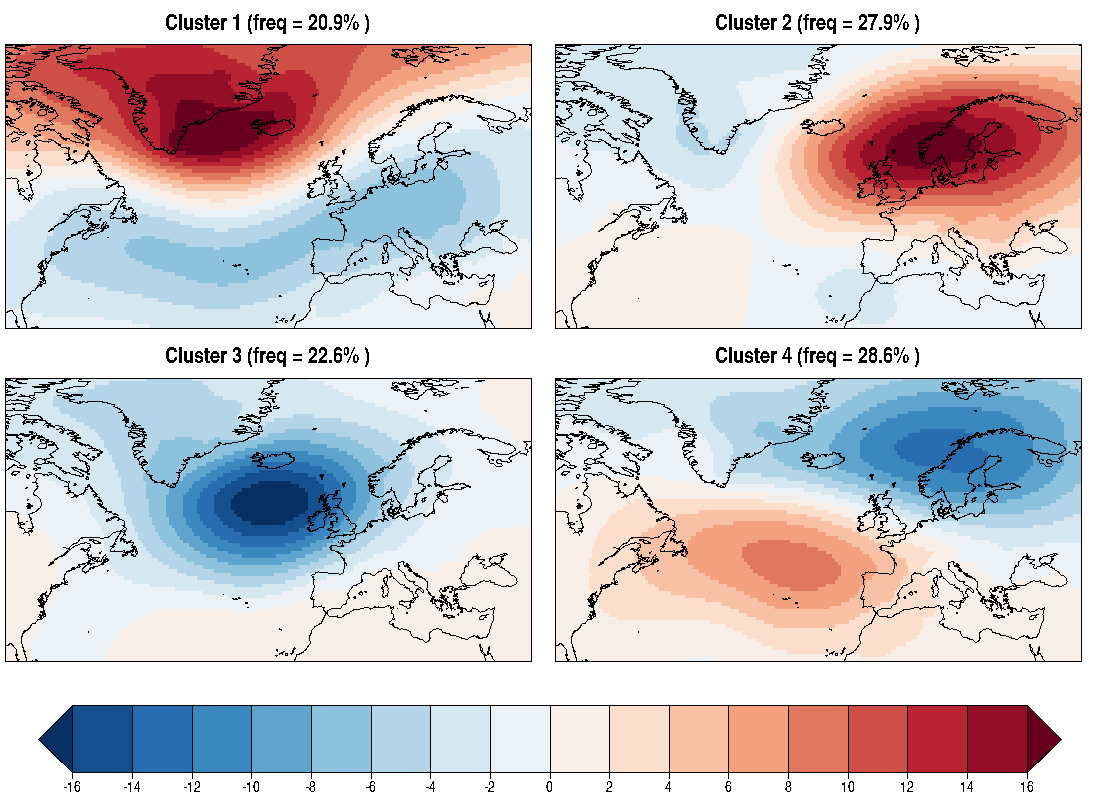
39.4 KB | W: | H:



