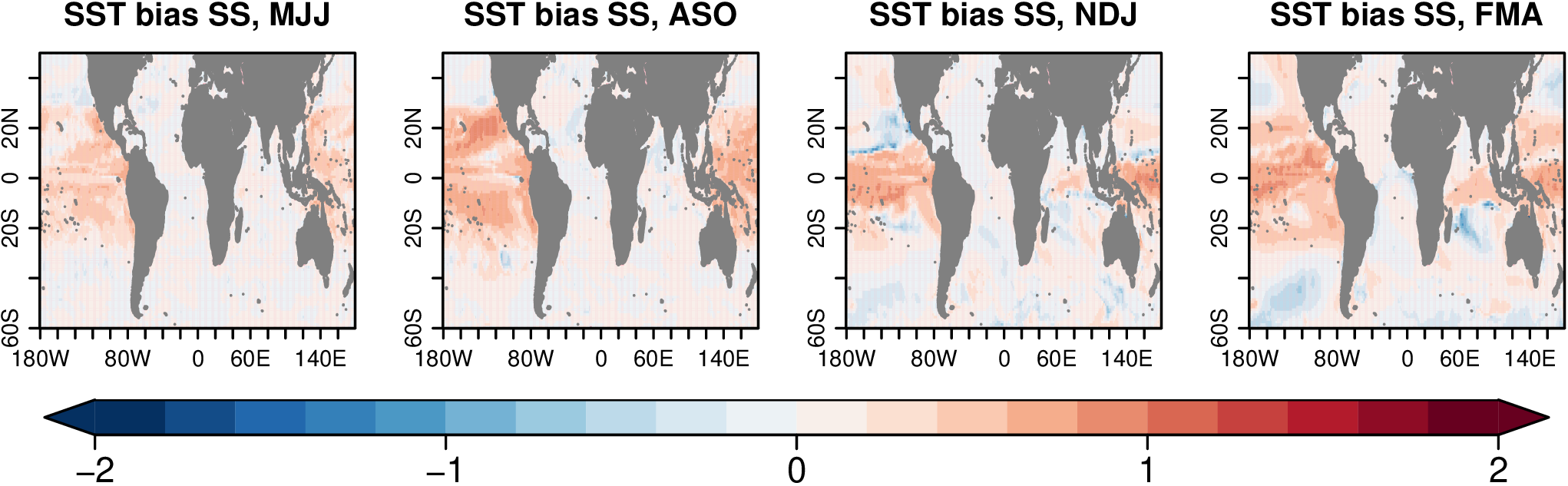
Abs Bias SS significance seems wrong
Hi @aho
I used the function AbsBiasSS but it does not seem to capture the significance. I am adding here the results, and when I have time I will add the plot where I calculate the significance with a bootstrap test to see the difference.
# Delete previous data
rm(list=ls())
gc()
library(boot)
library(s2dv)
library(abind)
library(s2dverification)
library(startR)
library(ncdf4)
library(multiApply)
library(SpecsVerification)
#source('AbsBiasSS.R')
#source('Bias.R')
#source('RandomWalkTest.R')
# ATL
lonmin <- -180
lonmax <- 180
latmin <- -60
latmax <- 50
sdates <- paste0(c(1981:2002), '0501')
# exp
repos_a511 <- paste0('/esarchive/scratch/eexarcho/Eleftheria/Pace_maker/Data/a511/tos/r360x180/',
'$var$_Omon_EC-Earth3-CC_troppac-pacemaker_s$sdate$-$member$_r360x180_$chunk$.nc')
repos_a53d <- paste0('/esarchive/scratch/eexarcho/Eleftheria/Pace_maker/Data/a53d/tos/r360x180/',
'$var$_Omon_EC-Earth3-CC_troppac-pacemaker_s$sdate$-$member$_r360x180_$chunk$.nc')
a511 <- Start(dat = repos_a511,
var = 'tos',
member = 'all',
sdate = sdates,
chunk = 'all',
# time = 'all',
time = indices(1:12), #first time step per day
chunk_depends = 'sdate',
time_across = 'chunk',
merge_across_dims = TRUE,
lat = values(list(latmin, latmax)),
lat_reorder = Sort(), ##1
lon = values(list(lonmin, lonmax)),
lon_reorder = CircularSort(-180, 180), ##2
synonims = list(lat = c('lat', 'latitude'),
lon = c('lon', 'longitude')),
return_vars = list(lon = 'dat', ##3
lat = 'dat', ##3
time = 'sdate'),
retrieve = T)
a53d <- Start(dat = repos_a53d,
var = 'tos',
member = 'all',
sdate = sdates,
chunk = 'all',
# time = 'all',
time = indices(1:12), #first time step per day
chunk_depends = 'sdate',
time_across = 'chunk',
merge_across_dims = TRUE,
lat = values(list(latmin, latmax)),
lat_reorder = Sort(), ##1
lon = values(list(lonmin, lonmax)),
lon_reorder = CircularSort(-180, 180), ##2
synonims = list(lat = c('lat', 'latitude'),
lon = c('lon', 'longitude')),
return_vars = list(lon = 'dat', ##3
lat = 'dat', ##3
time = 'sdate'),
retrieve = T)
###lons <- attr(exp, 'Variables')$common$tos$dim[[1]]$vals
###lats <- attr(exp, 'Variables')$common$tos$dim[[2]]$vals
lons <- as.vector(attr(a511, 'Variables')$dat1$lon)
lats <- as.vector(attr(a511, 'Variables')$dat1$lat)
dates <- attr(a511, 'Variables')$common$time
dates_file <- sort(unique(gsub('-', '', sapply(as.character(dates), substr, 1, 7))))
##
## obs
repos_obs <- '/esarchive/obs/ukmo/hadisst_v1.1/monthly_mean/$var$/$var$_$date$.nc'
obs.data <- Start(dat = repos_obs,
var = 'tos',
date = dates_file,
# time = 'all',
# time_across = 'date',
# merge_across_dims = TRUE,
# merge_across_dims_narm = TRUE,
## -----------------------------------------------------------------------
time = values(dates), #dim: [sdate = 2, time = 12]
#because time is assigned by 'values', set the tolerance to avoid too distinct match
time_var = 'time',
# time_tolerance = as.difftime(372, units = 'hours'),
#time values are across all the files
time_across = 'date',
#combine time and file_date dims
merge_across_dims = TRUE,
#exclude the additional NAs generated by merge_across_dims
merge_across_dims_narm = TRUE,
#split time dim, because it is two-dimensional
split_multiselected_dims = TRUE,
## -----------------------------------------------------------------------
lat = values(lats),
lon = values(lons),
#---------transform to get identical lat and lon------------ ##4
transform = CDORemapper,
transform_extra_cells = 2,
transform_params = list(grid = 'r360x180',
method = 'conservative',
crop = c(lonmin, lonmax, latmin, latmax)),
transform_vars = c('lat', 'lon'),
#-----------------------------------------------------------
synonims = list(lat = c('lat', 'latitude'),
lon = c('lon', 'longitude')),
return_vars = list(lat = NULL, ##3
lon = NULL, ##3
time = 'date'),
retrieve = TRUE)
abs.bias.a511.mjj <- s2dv::AbsBiasSS(a511.mjj, obs.mjj-273.15, a53d.mjj,
memb_dim = 'member', dat_dim='dat', ncores=16)
abs.bias.a511.aso <- s2dv::AbsBiasSS(a511.aso, obs.aso-273.15, a53d.aso,
memb_dim = 'member', dat_dim='dat', ncores=16)
abs.bias.a511.ndj <- s2dv::AbsBiasSS(a511.ndj, obs.ndj-273.15, a53d.ndj,
memb_dim = 'member', dat_dim='dat', ncores=16)
abs.bias.a511.fma <- s2dv::AbsBiasSS(a511.fma, obs.fma-273.15, a53d.fma,
memb_dim = 'member', dat_dim='dat', ncores=16)
min2=-2.
max2=2.
int2=(max2-min2)/20
interval2=seq(min2,max2,int2)
clim.palette(palette = "bluered")
color=clim.colors(20, palette = "bluered")
pdf( "../Figures/SST_bias_SS_a511_a53d.pdf" )
nf <- layout(matrix(c(1, 2 , 3, 4, 5, 5, 5, 5),2,4,byrow=TRUE),
widths=1, heights=c(1,0.3), TRUE)
s2dv::PlotEquiMap( abs.bias.a511.mjj$biasSS[1, 1, 1, 1, , ],
lons, lats ,
toptitle= "SST bias SS, MJJ",
cols=color, brks=interval2, drawleg = F,
triangle_ends = c(T,T), col_inf = color[1], col_sup = color[20],
filled.continents = T,
dots = abs.bias.a511.mjj$sign[1, 1, 1, 1, ,],
dot_size = 0.5,
numbfig=4
)
s2dv::PlotEquiMap( abs.bias.a511.aso$biasSS[1, 1, 1, 1, , ],
lons, lats ,
toptitle= "SST bias SS, ASO",
cols=color, brks=interval2, drawleg = F,
triangle_ends = c(T,T), col_inf = color[1], col_sup = color[20],
filled.continents = T,
dots = abs.bias.a511.aso$sign[1, 1, 1, 1, ,],
dot_size = 0.5,
numbfig=4
)
s2dv::PlotEquiMap( abs.bias.a511.ndj$biasSS[1, 1, 1, 1, , ],
lons, lats ,
toptitle= "SST bias SS, NDJ",
cols=color, brks=interval2, drawleg = F,
triangle_ends = c(T,T), col_inf = color[1], col_sup = color[20],
filled.continents = T,
dots = abs.bias.a511.ndj$sign[1, 1, 1, 1, ,],
dot_size = 0.5,
numbfig=4
)
s2dv::PlotEquiMap( abs.bias.a511.fma$biasSS[1, 1, 1, 1, , ],
lons, lats ,
toptitle= "SST bias SS, FMA",
cols=color, brks=interval2, drawleg = F,
triangle_ends = c(T,T), col_inf = color[1], col_sup = color[20],
filled.continents = T,
dots = abs.bias.a511.fma$sign[1, 1, 1, 1, ,],
dot_size = 0.5,
numbfig=4
)
ColorBar(brks=interval2,
triangle_ends = c(T,T), col_inf = color[1], col_sup = color[20],
cols = color, vert = FALSE)
dev.off()
#